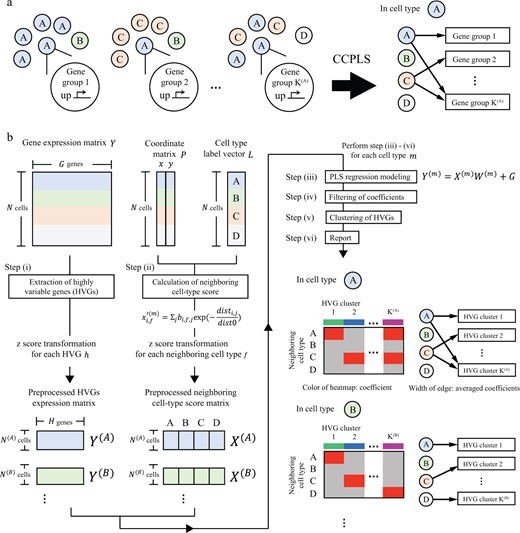
・We propose CCPLS, which is a statistical framework for identifying cell–cell communications as the effects of multiple neighboring cell types on cell-to-cell expression variability of highly variable genes (HVGs), based on the spatial transcriptome data.
・For each cell type, CCPLS performs PLS regression modeling and reports coefficients as the quantitative index of the cell–cell communications. Evaluation using simulated data showed our method accurately estimated the effects of multiple neighboring cell types on HVGs.
・Furthermore, applications to the two real datasets demonstrate that CCPLS can extract biologically interpretable insights from the inferred cell–cell communications.
-
最近の投稿
アーカイブ
カテゴリー